Code
library(reticulate)
library(ggplot2)
theme_set(theme_classic())library(reticulate)
library(ggplot2)
theme_set(theme_classic())import mne
import os
import pandas as pd
from plotnine import *
from matplotlib import pyplot as plt
from glob import glob
import pickle
import numpy as np
from scipy.interpolate import CubicSpline, PchipInterpolator
from statsmodels import api as sm
from scipy.signal import unit_impulse
from scipy.optimize import curve_fittheme_set(theme_classic()) # ggplot
Fs=500 # eyetracking data was sampled at 500Hz
ds = 'nback'
instr_onsets = 4.4 + (3.6 + 1.2 + 24) * np.arange(12)
blank_onsets = instr_onsets + 3.6
block_onsets = blank_onsets + 1.2
stim_onsets = (block_onsets[:, np.newaxis] + 2.4 * np.arange(10)).flatten()#ds = 'data/ET/*s*r*nBack*.asc' if ds == 'nback' else 'data/ET/*s1*r*cardWager*.asc'
#all_data = [*map(mne.io.read_raw_eyelink, glob(ds))]
# Don't forget to change! vv
with open('data_nback_noproc.pkl', 'rb') as file:
all_data = pickle.load(file)
# pickle.dump(all_data, file)def crop(raw):
events, event_ids = mne.events_from_annotations(
raw, regexp='RUN (OFFSET|ONSET TRIGGER)'
)
return raw.copy().crop(*(events[:, 0] / raw.info['sfreq']))
all_data_cropped = [data for data in map(crop, all_data) if data.times[-1] > 359]Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']
Used Annotations descriptions: ['RUN OFFSET', 'RUN ONSET TRIGGER']d = len(all_data) - len(all_data_cropped)
if d > 0:
print('%d file(s) excluded due to insufficient data' % d)1 file(s) excluded due to insufficient datapupil = all_data_cropped[0].get_data()[2,:]
# The following article supports use of a method other than Cubic Spline
# for interpolation:
#
# Dan, E. L., Dînşoreanu, M., & Mureşan, R. C. (2020). Accuracy of Six
# Interpolation Methods Applied on Pupil Diameter Data. 2020 IEEE International
# Conference on Automation, Quality and Testing, Robotics (AQTR), 1–5.
# https://doi.org/10.1109/AQTR49680.2020.9129915
#
# I've thus chosen to use the Piecewise Cubic Hermite Interpolating Polynomial method.
def interpolate(signal, buffer=(250, 50), method='pchip'):
sig = signal.copy()
for i, v in enumerate(sig):
if v <= 0 or np.isnan(v):
signal[max(0, i-buffer[0]):min(i+buffer[1], len(signal))] = np.nan
iint = np.isnan(signal)
sample_index = np.arange(len(signal))
match method:
case 'pchip':
pchip = PchipInterpolator(
sample_index[~iint],
signal[~iint],
extrapolate=False
)
signal[iint] = pchip(sample_index[iint])
case 'cs':
csi = CubicSpline(
sample_index[~iint],
signal[~iint],
bc_type='natural',
extrapolate=False
)
signal[iint] = csi(sample_index[iint])
return signal
z = pupil.copy()
z = interpolate(z, method='pchip')
z -= z.mean(where=~np.isnan(z))
z /= np.sqrt(z.var(where=~np.isnan(z)))
plt.figure(figsize=(18, 4))
plt.vlines(block_onsets, ymin=-2.5, ymax=2.5, color='y')
plt.vlines(instr_onsets, ymin=-2.5, ymax=2.5, color='g')
plt.vlines(blank_onsets, ymin=-2.5, ymax=2.5, color='b')
plt.plot(np.arange(len(z)) / Fs, z, 'r')
plt.ylim(-4,4)(-4.0, 4.0)plt.show()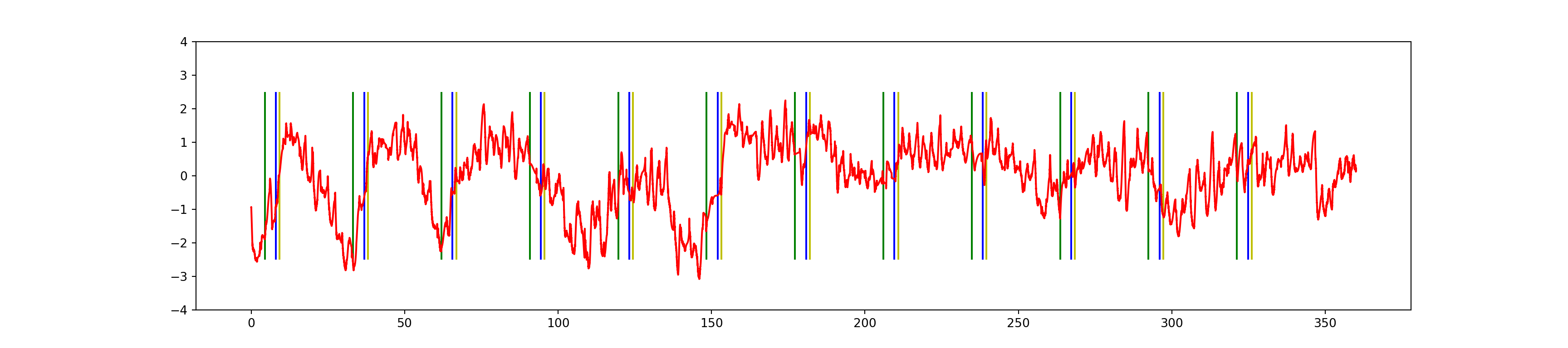
colors = {
'blank': 'lightgrey',
'instr': 'lightblue',
'block': 'yellow',
'stim': 'grey'
}
z = pupil.copy()
z = interpolate(z, method='cs')
z -= z.mean(where=~np.isnan(z))
z /= np.sqrt(z.var(where=~np.isnan(z)))
w = pupil.copy()
w = interpolate(w, method='pchip')
w -= w.mean(where=~np.isnan(w))
w /= np.sqrt(w.var(where=~np.isnan(w)))
d = pd.concat([
pd.DataFrame({'t': np.arange(len(z))/Fs, 'data': z, 'method': 'CS'}),
pd.DataFrame({'t': np.arange(len(w))/Fs, 'data': w, 'method': 'PCHIP'})
])@fig-proc-interp-compare compares interpolation methods
ggplot(py$d) +
geom_line(aes(t, data), color='red', size=1) +
facet_grid(method~.)Warning in py_to_r.pandas.core.frame.DataFrame(x): index contains duplicated
values: row names not setWarning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.x = (
ggplot()
# blanks
+ geom_rect(aes(xmin=np.concatenate(([0], blank_onsets, [360-10])), xmax=np.concatenate(([[instr_onsets[0]], block_onsets, [360]]))), ymax=6, ymin=-6, fill=colors['blank'])
# instructions
+ geom_rect(aes(xmin=instr_onsets, xmax=blank_onsets), ymax=6, ymin=-6, fill=colors['instr'])
# blocks
+ geom_rect(aes(xmin=block_onsets, xmax=block_onsets+24), ymax=6, ymin=-6, fill=colors['block'])
# stimulus onsets
+ geom_vline(aes(xintercept=stim_onsets), color='grey', alpha=0.2)
# signal
+ geom_line(aes('t', 'data'), data=d.query('method == "PCHIP"'), color='red', size=1)
+ labs(title='Piecewise Cubic Hermite Interpolating Polynomial', y='Pupil Size (SD)', x='Time (s)')
# + geom_line(aes('t', 'data'), data=d.query('method == "CS"'), color='red', size=1)
# + labs(title='Cubic Spline Interpolation', y='Pupil Size (SD)', x='Time (s)')
+ scale_x_continuous(breaks=np.concatenate([[0], block_onsets, [360]]), expand=(0,0))
+ scale_y_continuous(limits=(-6,6), expand=(0,0))
+ theme(figure_size=(18,5))
);
# ggsave(x, 'interp-cspline.png')
# ggsave(x, 'interp-pchip.pdf')
x.show();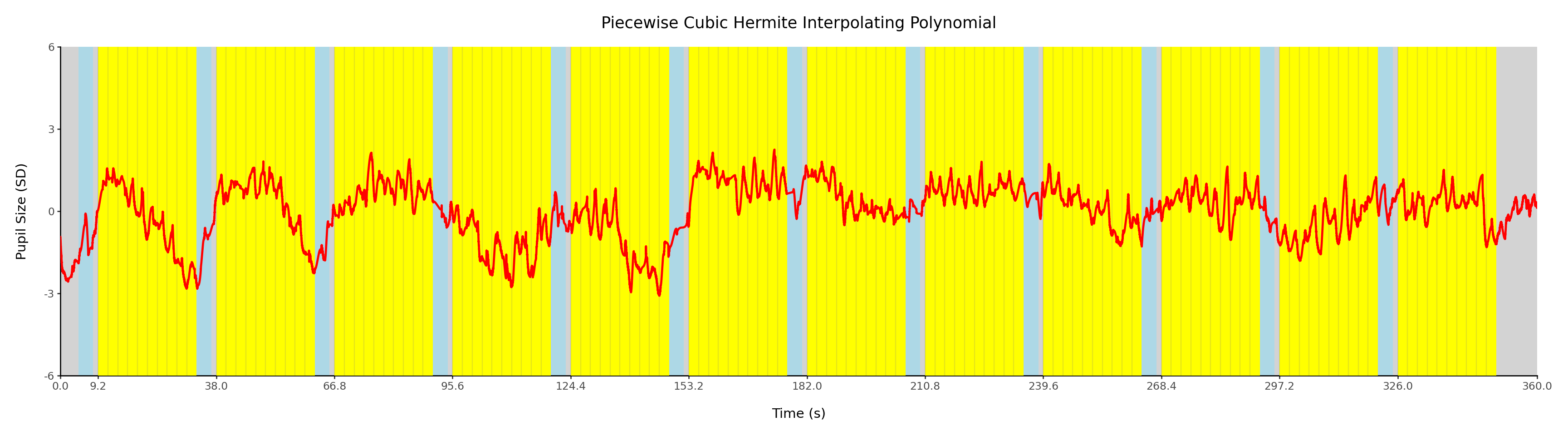
Per @fig-proc-interp-compare we chose PCHIP as our interpolation method.
all_data_preproc = []
files = []
for i, ds in enumerate(all_data_cropped):
x, y, p = ds.get_data()
xc = x[~np.isnan(x)]
if len(xc) < .80 * len(x):
print('More than 20%% x data missing (%.3f seconds) for dataset %d.' %
((len(x)-len(xc))/Fs, i)
)
continue
xci = xc[(xc > 200) & (xc < 800)]
if len(xci) < .8*len(x):
print('More than 20%% x data out of bounds (%.3f seconds) for dataset %d.' %
((len(x)-len(xci))/Fs, i),
(xc.mean() + np.array([-1, 1]) *np.sqrt(xc.var())).astype(int)
)
continue
yc = y[~np.isnan(y)]
if len(yc) < .80 * len(y):
print('More than 20%% y data missing (%.3f seconds) for dataset %d.' %
((len(y)-len(yc))/Fs, i)
)
continue
yci = yc[(yc > 200) & (yc < 1000)]
if len(yci) < .8*len(y):
print('More than 20%% y data out of bounds (%.3f seconds) for dataset %d.' %
((len(y)-len(yci))/Fs, i),
(yc.mean() + np.array([-1, 1]) *np.sqrt(yc.var())).astype(int)
)
continue
if p.sum() < 1:
print('Pupil empty for dataset %d' % i)
print(ds)
continue
try:
p = interpolate(p, method='pchip')
except:
print('Error processing dataset %d' % i)
print(ds)
continue
p -= p.mean(where=~np.isnan(p))
p /= np.sqrt(p.var(where=~np.isnan(p)))
# Ensure all arrays are the same size.
if len(p) > 180000:
p = p[:180000] # crop to 360s * 500Hz
elif len(p) < 180000:
p = np.concatenate([p, np.empty(180000 - len(p))]) # pad ends with NaN
all_data_preproc.append(p)
files.append(ds.filenames[0].as_posix())More than 20% x data out of bounds (352.740 seconds) for dataset 1. [1152 1628]
More than 20% x data out of bounds (77.322 seconds) for dataset 2. [331 787]
More than 20% x data missing (76.316 seconds) for dataset 13.
More than 20% x data missing (101.294 seconds) for dataset 15.
More than 20% x data missing (74.540 seconds) for dataset 25.
More than 20% x data missing (360.004 seconds) for dataset 29.
More than 20% x data missing (358.878 seconds) for dataset 30.
More than 20% x data out of bounds (154.970 seconds) for dataset 33. [ 307 1009]
More than 20% x data missing (81.950 seconds) for dataset 34.
More than 20% x data missing (99.324 seconds) for dataset 38.
More than 20% x data missing (359.004 seconds) for dataset 39.
More than 20% x data out of bounds (83.530 seconds) for dataset 40. [466 716]
More than 20% x data out of bounds (73.930 seconds) for dataset 41. [424 559]
More than 20% x data out of bounds (104.542 seconds) for dataset 43. [236 668]
More than 20% x data missing (99.886 seconds) for dataset 45.
More than 20% x data missing (73.226 seconds) for dataset 46.
More than 20% x data out of bounds (77.472 seconds) for dataset 48. [281 685]
More than 20% x data out of bounds (351.238 seconds) for dataset 49. [1105 1662]
More than 20% x data missing (351.428 seconds) for dataset 51.
More than 20% y data out of bounds (349.792 seconds) for dataset 53. [-46 140]
More than 20% x data out of bounds (79.060 seconds) for dataset 61. [426 570]
More than 20% y data out of bounds (350.146 seconds) for dataset 62. [-110 93]
More than 20% x data missing (89.162 seconds) for dataset 64.
More than 20% x data missing (73.544 seconds) for dataset 66.
More than 20% x data missing (72.056 seconds) for dataset 69.
More than 20% x data missing (80.648 seconds) for dataset 71.
More than 20% x data out of bounds (96.342 seconds) for dataset 72. [291 839]
More than 20% x data missing (360.006 seconds) for dataset 78.
More than 20% x data out of bounds (95.164 seconds) for dataset 82. [269 661]
More than 20% x data out of bounds (154.680 seconds) for dataset 84. [ 32 565]
d = len(all_data_cropped) - len(all_data_preproc)
if d > 0:
print('%d file(s) excluded due to missing/out of bounds data' % d)30 file(s) excluded due to missing/out of bounds data
data = np.column_stack(all_data_preproc)behavioral = 'data/BH'
behfiles = [
os.path.join(
behavioral,
os.path.basename(file[:-4]).removeprefix('eyeTrack_') + '.csv'
) for file in files
]
order = np.column_stack([
pd.read_csv(file)['BlockType'][10 * np.arange(12)].to_numpy() for file in behfiles
])
order[:,:4] #sample the first four columns, order will have 12 rows (one for each block) describing the block order for that session.array([['1back', '2back', '2back', '1back'],
['0back', '1back', '1back', '0back'],
['2back', '0back', '0back', '2back'],
['0back', '1back', '1back', '0back'],
['1back', '2back', '2back', '1back'],
['2back', '0back', '0back', '2back'],
['1back', '2back', '2back', '1back'],
['2back', '0back', '0back', '2back'],
['0back', '1back', '1back', '0back'],
['2back', '0back', '0back', '2back'],
['1back', '2back', '2back', '1back'],
['0back', '1back', '1back', '0back']], dtype=object)a = np.mean(data, axis=1, where=~np.isnan(data))
x = (
ggplot()
# blanks
+ geom_rect(aes(xmin=np.concatenate(([0], blank_onsets, [360-10])), xmax=np.concatenate(([[instr_onsets[0]], block_onsets, [360]]))), ymax=6, ymin=-6, fill=colors['blank'])
# instructions
+ geom_rect(aes(xmin=instr_onsets, xmax=blank_onsets), ymax=6, ymin=-6, fill=colors['instr'])
# blocks
+ geom_rect(aes(xmin=block_onsets, xmax=block_onsets+24), ymax=6, ymin=-6, fill=colors['block'])
# stimulus onsets
+ geom_vline(aes(xintercept=stim_onsets), color='grey', alpha=0.2)
# signal
+ geom_line(aes(np.arange(180000)/Fs, a), color='red', size=1)
+ labs(title='Mean Signal, 58 Sessions', y='Pupil Size (SD)', x='Time (s)')
+ scale_x_continuous(breaks=np.concatenate([[0], block_onsets, [360]]), expand=(0,0))
+ scale_y_continuous(limits=(-2,2), expand=(0,0))
+ theme(figure_size=(18,5))
)
x.show()/usr3/graduate/mpascale/.local/lib/python3.10/site-packages/plotnine/geoms/geom_path.py:100: PlotnineWarning: geom_path: Removed 1 rows containing missing values.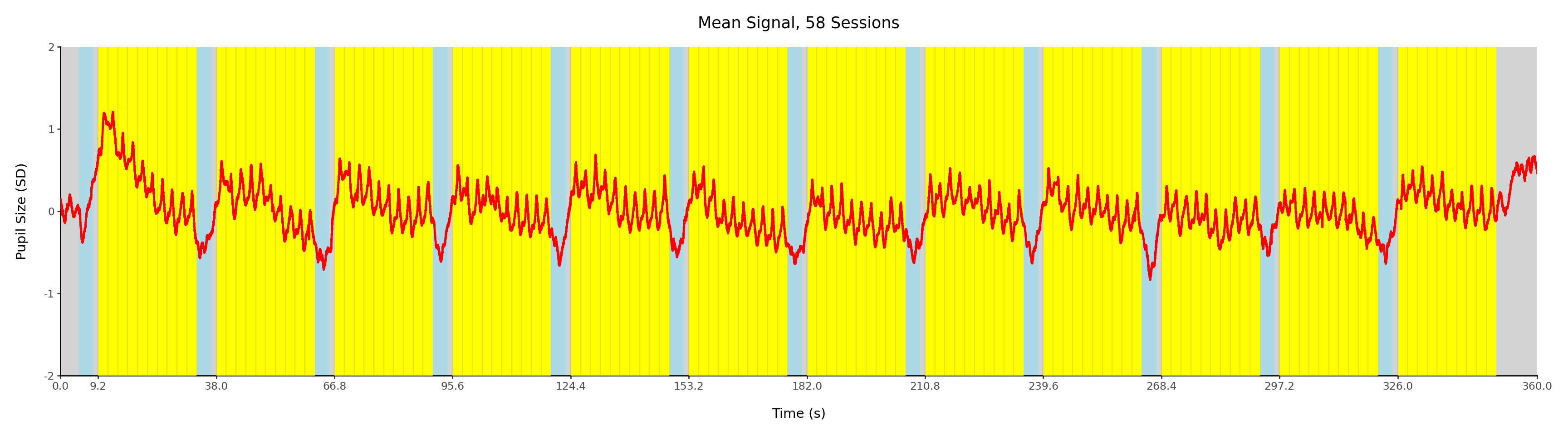
# Define the canonical pupillary response function
purf = (lambda n=10.1, tmax=0.93 * Fs: (lambda t: np.where(t > 0, np.power(t, n) * np.exp(-n * t / tmax), 0)))
x = np.linspace(0,2.5*Fs,100)
p = purf()(x)
p /= p.max()
x = ggplot() + geom_line(aes(x*1000/Fs,p), color='red', size=1) + geom_vline(xintercept=930, linetype='dashed') + labs(y='Pupil Response', x='Time (ms)', title='Canonical PuRF')
x.show()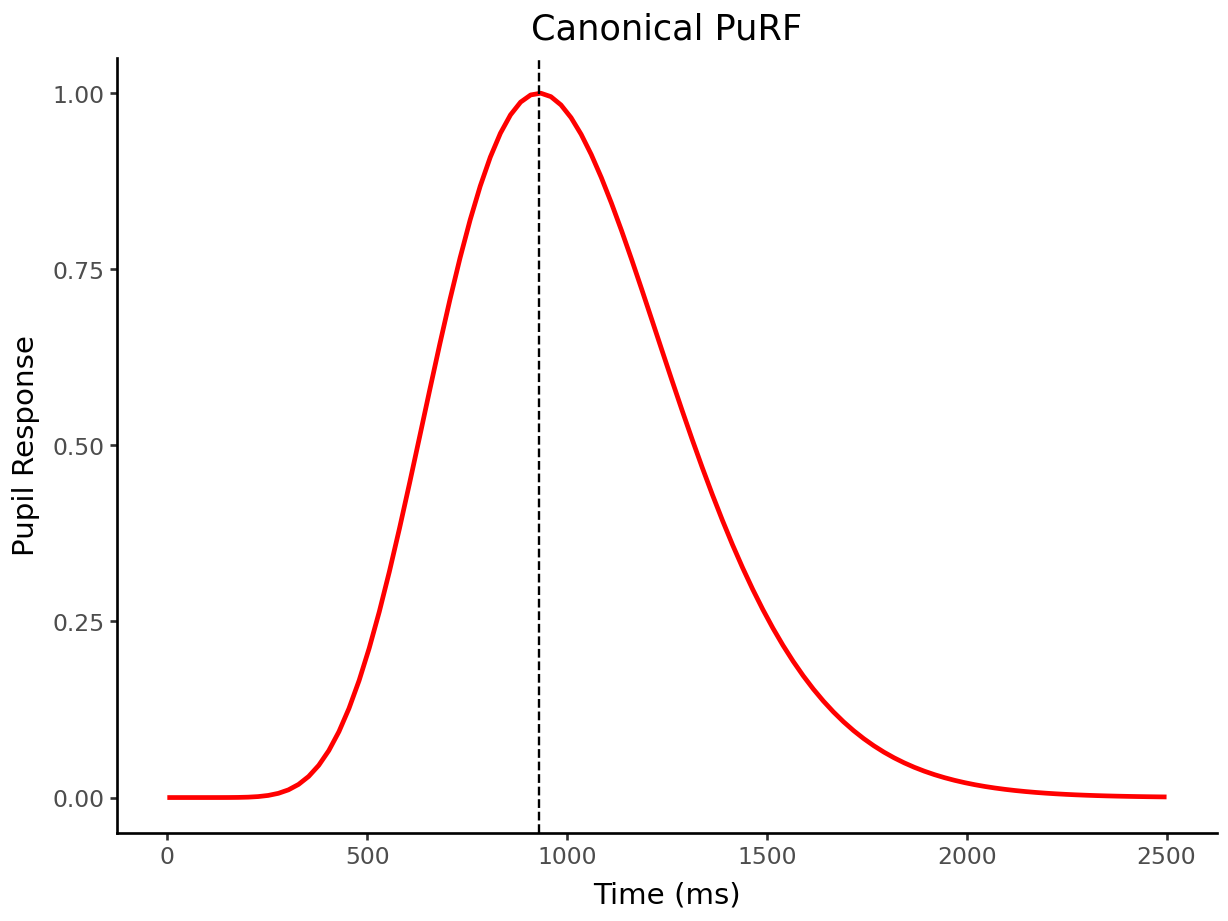
# Create a hypothetical response expected during the task, according to:
# Denison, R. N., Parker, J. A., & Carrasco, M. (2020). Modeling pupil
# responses to rapid sequential events. Behavior Research Methods, 52(5),
# 1991–2007. https://doi.org/10.3758/s13428-020-01368-6
# A square signal at low amplitude, simple effect of the task itself.
def boxcar(T, ts, te):
assert ts < te, "Boxcar start time must be less than end time."
box = np.zeros(T)
box[ts:te] = 1
return box
# A linear combination of the boxcar task signal, two impulses associated with the
# stimulus and some internal event.
def hypo_curve(x, tmax, imp1_t, imp1_amp, imp2_t, imp2_amp, box_ts, box_te, box_amp):
T = len(x)
# Canonincal PuRF
p = purf(tmax=tmax)(x)
# Input Vectors (Impulses)
imps = [unit_impulse(T, imp1_t),
unit_impulse(T, imp2_t),
boxcar(T, box_ts, box_te)]
amps = [imp1_amp, imp2_amp, box_amp]
# Convolved PuRF for each impulse
cnvs = [np.convolve(p, i, 'full') for i in imps]
# Normalize the convolved signals and multiply by amplitude
cnvs = [(x / max(x)) * amps[i] for i, x in enumerate(cnvs)]
# Return the sum (linear combination) of the convolutions
comb = np.sum(cnvs, axis=0)
# comb = comb / comb.max()
return comb, cnvs, imps
x = np.arange(0,2.5*Fs)
# p = purf()(x)
# p /= p.max()
# x = ggplot() + geom_line(aes(x*1000,p), color='red', size=1) + geom_vline(xintercept=930, linetype='dashed') + labs(y='Pupil Response', x='Time (ms)', title='Canonical PuRF')
# ggsave(x, 'purf.png')
# x
c, cnvs, imps = hypo_curve(x, .5*Fs, 0, 2, Fs, 6, 0, int(2.4*Fs), 1)
plt.plot(c/c.max(), 'orange')
for ci in cnvs:
plt.plot(ci/c.max(), 'grey')
plt.title('PuRF Model - Convolved Signals')
plt.xlabel('Time (s)')
plt.ylabel('Pupil Response')
plt.show()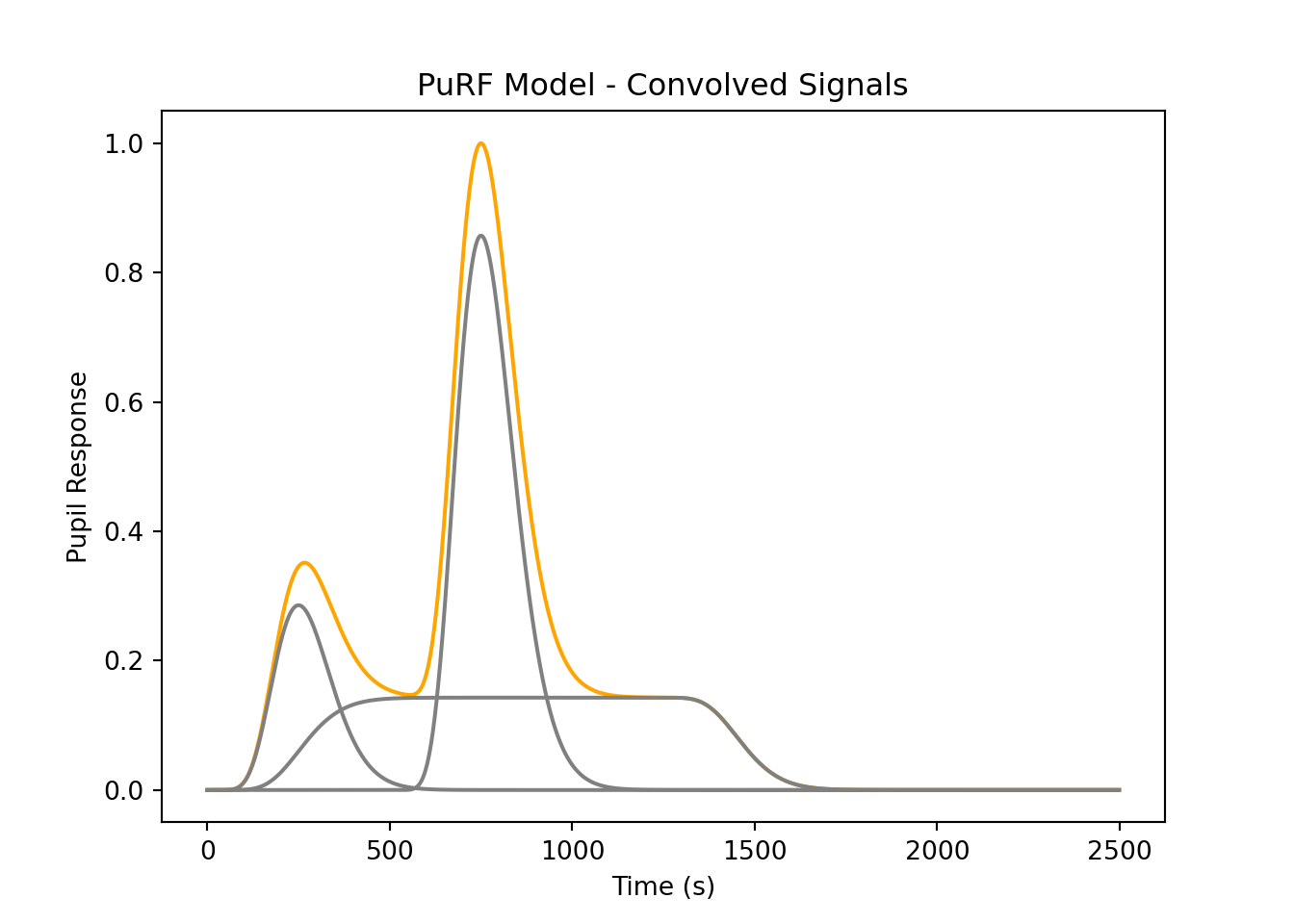
# Create Y, the dependent variable
# Remove the 4.2s start blank and 10s end blank
Y = data[int(4.2 * Fs):int(180000-10*Fs),:]
k = np.arange(int(4.8*Fs), int((24+4.8)*Fs))[:,np.newaxis] + ((24 + 4.8) * Fs * np.arange(12)).astype(int)
Y = Y[k.flatten('F'),:]
Ysh = Y.shape
plt.plot(Y[:, 0])
Y = Y.flatten('F')
# all data in single column
plt.matshow(Y[:,np.newaxis], aspect='auto')
Ysh[0]144000
# Create X, the predictors/covariates
# First, predictors for nback block
X = np.column_stack([np.repeat((order == bl).astype(int).flatten(), 24*Fs) for bl in np.unique(order)])
# Intercept
X = sm.add_constant(X)
# Regressors for low frequency drift
def drift_regressors_cosine(f, samples):
assert type(f) is int and f > 0, "Frequency must be a nonzero integer."
x = np.linspace(0,1,samples)
return np.column_stack([np.cos(np.pi * i * x) for i in np.arange(f)+1])
cossdrift = drift_regressors_cosine(4,Ysh[0])
X = np.hstack([X, np.vstack([cossdrift]* 58)])
X = np.hstack([X, np.column_stack([np.tile(c[:int(2.4*Fs)], 58*10*12)])])
plt.matshow(X[:,0:(24*12*Fs)], aspect='auto')
plt.show()
model = sm.regression.linear_model.OLS(Y, X)
fit = model.fit()
fit.summary() # This is not fitting but we haven't adjusted to PuRF signal to each subjects as done by the authors. Namely, the last coefficient is miniscule.| Dep. Variable: | y | R-squared: | 0.023 |
| Model: | OLS | Adj. R-squared: | 0.023 |
| Method: | Least Squares | F-statistic: | 2.505e+04 |
| Date: | Tue, 04 Jun 2024 | Prob (F-statistic): | 0.00 |
| Time: | 12:15:58 | Log-Likelihood: | -1.1708e+07 |
| No. Observations: | 8352000 | AIC: | 2.342e+07 |
| Df Residuals: | 8351991 | BIC: | 2.342e+07 |
| Df Model: | 8 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
| const | 6.843e+05 | 6.88e+06 | 0.099 | 0.921 | -1.28e+07 | 1.42e+07 |
| x1 | -6.843e+05 | 6.88e+06 | -0.099 | 0.921 | -1.42e+07 | 1.28e+07 |
| x2 | -6.843e+05 | 6.88e+06 | -0.099 | 0.921 | -1.42e+07 | 1.28e+07 |
| x3 | -6.843e+05 | 6.88e+06 | -0.099 | 0.921 | -1.42e+07 | 1.28e+07 |
| x4 | 0.1010 | 0.000 | 209.761 | 0.000 | 0.100 | 0.102 |
| x5 | 0.0923 | 0.000 | 191.884 | 0.000 | 0.091 | 0.093 |
| x6 | 0.0372 | 0.000 | 76.911 | 0.000 | 0.036 | 0.038 |
| x7 | 0.0449 | 0.000 | 93.361 | 0.000 | 0.044 | 0.046 |
| x8 | 0.0549 | 0.000 | 294.140 | 0.000 | 0.054 | 0.055 |
| Omnibus: | 41675.076 | Durbin-Watson: | 0.001 |
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 60078.739 |
| Skew: | -0.010 | Prob(JB): | 0.00 |
| Kurtosis: | 3.415 | Cond. No. | 1.18e+11 |
plt.figure(figsize=(18,5))
plt.plot(Y[0:(24*12*Fs)])
plt.plot(fit.predict(X[0:(24*12*Fs),:])) # Just looking at a section of predicted signal. This kinda looks like it's picking up on only the block type.
plt.plot(Y)
plt.plot(fit.predict()) # Just looking at a section of predicted signal. This kinda looks like it's picking up on only the block type.